Prokaryotes with a nucleus?
Back to main indexIn fact, it is possible to find eukaryotic characteristics in prokaryotes: We note the presence of a primitive cytoskeleton in eubacteria thanks to the FtsZ protein which is a bacterial homolog of the eukaroytic protein tubulin. Likewise, the MreB protein assembles into filaments and has a 3D structure similar to actin. Two genes encoding true α and β-type tubulins have been discovered in Prosthecobacter, which are aerobic bacteria from the Verrucomicrobia group, others have been detected in some archaea. The data concerning the origin of these tubulins are contradictory: the comparison of prokaryotic sequences with eukaryotic proteins shows large discrepancies, indicating that they would probably not result from a horizontal transfer between these domains of living things. However, the presence of these genes only in certain strains of Prosthecobacter or archaea, but not in others, would on the contrary be indicative of a horizontal transfer followed by an accelerated evolution. More recently, proteins clearly orthologous to actin have been identified in archaea. Called “crenactins”, these proteins polymerize to form helical structures that cross cells and which probably participate in the maintenance of shape and cell division. Another characteristic of eukaryotes is also found in several prokaryotes: the complexity of the genetic material. Indeed, we sometimes note the presence of several chromosomes, some of which are linear. In some archaea, histones structure the chromatin into nucleosomes. Introns, long considered to be specific to eukaryotes, are present in many species of archaea.
However, the internal structuring of the cell by a network of organelles is only found in a small number of eubacterial groups. These internal membrane systems often make it possible to compartmentalize specific enzymatic reactions: thylakoids of cyanobacteria where photosynthesis occurs, internal membrane systems of endobiont sponge bacteria (Poribacteria), vesicles of methylotrophic bacteria or “anammoxosomes”, the seat of a anaerobic oxidation of ammonium in Planctomycetales. These Planctomycetales (or Planctomycetes) have many interesting properties bringing them closer to eukaryotes or archaea. They have no peptidoglycans and divide by budding. Species denitrifying in the absence of oxygen have the ability to synthesize ether-type lipids which is a characteristic of archaea! Others have the ability to synthesize sterols like eukaryotes! In some, such as Pirellula marina and Pirellula staleyi, heterotrophic planctomycetales found in the Baltic Sea, the DNA is surrounded by a single lipid membrane: the intracytoplasmic membrane. This structure is called a pirellulosome (Figure 11) bathed in the paryphoplasm. RNA is found in both compartments.
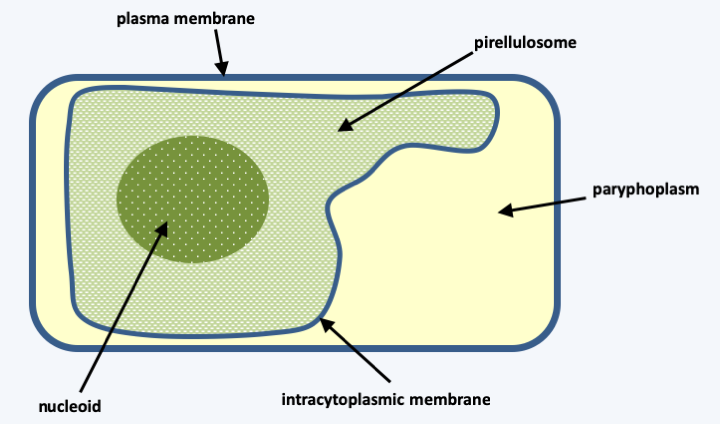
Figure 11.
The pirellulosome is surrounded by a single membrane defining two internal compartments. The nucleoid is located in the pirellulosome which is surrounded by the paryphoplasm.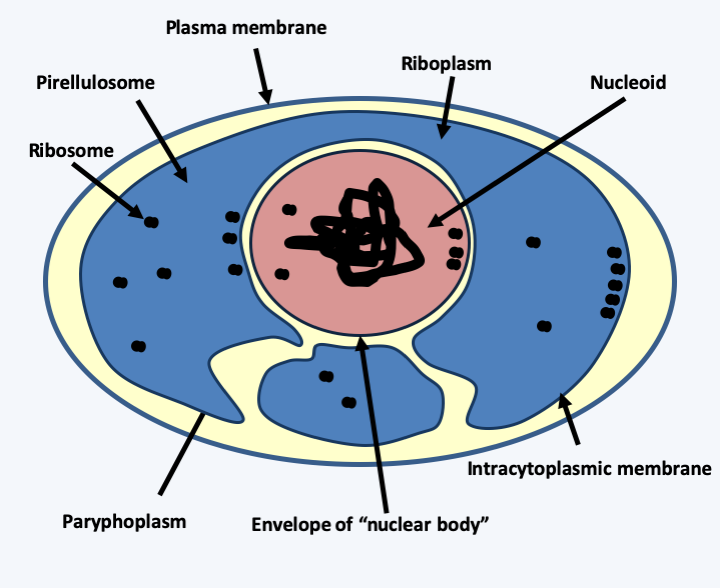
Figure 12.
The structure of Gemmata obscuriglobus. The pirellulosome is more or less lobed.The most surprising species of planctomycetes is undoubtedly Gemmata obscuriglobus, a bacterium that grows in freshwater ponds in Australia. It has several unorthodox characteristics for a bacterium, including:
- The ability to synthesize sterols.
- A genome surrounded first by a pirellulosome and by another lipid membrane, the two being separated by an inter-membrane space; the latter structure therefore strongly resembles a nuclear double membrane which would delimit “a nuclear body” (Figure 12). For the moment, it is suspected that this membrane is the result of convergent evolution and it is not known whether it causes the decoupling between transcription and translation observed in eukaryotes. However, it seems that RNA is present in both compartments, suggesting a possible decoupling for some mRNAs.
- Cell division with specific characteristics that are not found in eukaryotes or in other bacteria (Figure 13).
- Proteins resembling eukaryotic proteins which cover the vesicles of exo/endocytosis (coat-proteins). The divergence between these bacterial proteins and those of eukaryotes is too great to make molecular phylogenies. Nevertheless, the organization of the different domains is conserved, suggesting that prokaryotic and eukaryotic proteins could be related. Gemmata obscuriglobus is not the only species in the “Planctomycetes - Verrumicrobia - Chlamydiae” clade that has these proteins, but it is the one that is the most studied. One of these proteins has been shown to be localized in part in association with intracytoplasmic vesicles. There also seems to be a phenomenon related to endocytosis, even if for these bacteria it is highly probable that this capacity appeared by evolutionary convergence. Indeed, Gemmata obscuriglobus is able to import proteins from the external environment via vesicles resembling endocytosis vesicles. This can be seen simply by incubating the bacteria in a solution containing GFP (green fluorescent protein). Quickly, the cytoplasm of the bacteria becomes fluorescent. This property does not seem to be restricted to Gemmata obscuriglobus because recently it has been shown that photosynthetic bacteria (Rhodobacter sphaeroides) are also capable of producing vesicles by budding from the plasma membrane.
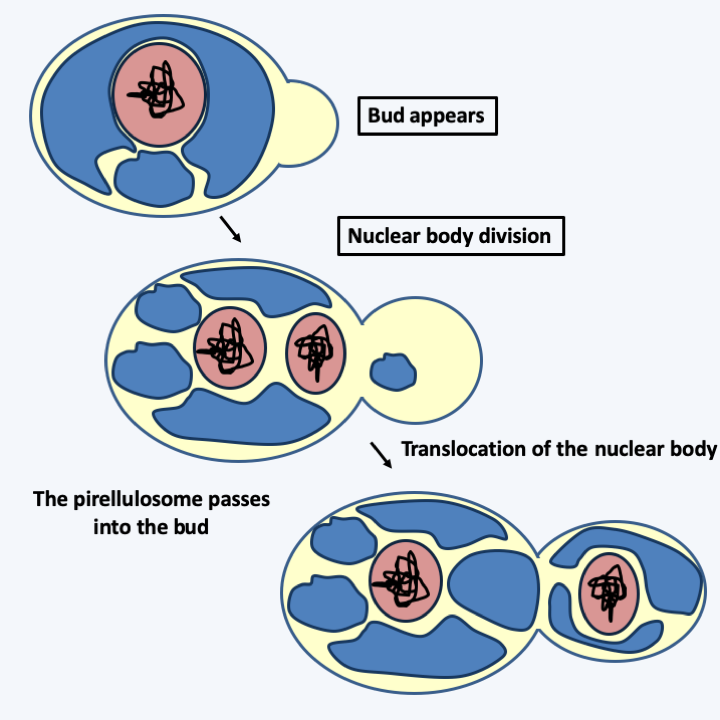
Figure 13.
Model of cell division of Gemmata obscuriglobus.Despite this set of data comparing Gemmata obscuriglobus to eukaryotes, analysis of 16S and 5S ribosomal RNA sequences shows that it is indeed from an eubacterium which is located at the base of the phylogenetic tree. In particular, diphtheria toxin is unable to catalyze the covalent binding of adenosine diphosphate ribose (ADPR) to the translational elongation factor EF2 (= EF-G) of Gemmata obscuriglobus. This is a property specific to eubacteria because the toxin effectively inactivates the aEF2 and eEF2 proteins of archaea and eukaryotes. In addition, the genome of this bacterium is now completely sequenced and does not show an accumulation of genes that would resemble those of eukaryotes more than those of other bacteria. On the other hand, these organisms have relics of genes involved in the synthesis of peptidoglycan, showing that they are derived from more classical bacteria. All of these data show that Gemmata obscuriglobus is actually a bacterium that evolved convergently with eukaryotes. It does not appear to be the eukaryotic “ancestor”, but gives an idea of what this ancestor may have looked like.
In conclusion, these examples show that most of the characteristics of eukaryotes are found in a simpler form in prokaryotes, including the compartmentalization of DNA in membrane structures. The differences between the two cell types are not qualitative but rather quantitative. As for the bacteria described in this chapter, despite the presence of a nucleoid surrounded by a membrane, these organisms are true prokaryotes and eukaryotes are unlikely to derive from them. Nevertheless, the group of Planctomycetales/Verrucomicrobia exhibits many interesting characteristics and it is possible that in the near future an organism related to the ancestor of eukaryotes will be discovered in this group. Currently no bacteria have a true network of internal membranes homologous to the nucleus/reticulum/golgi network. In fact, no prokaryote seems to be able to perform phagocytosis, which therefore remains a eukaryotic characteristic. Nevertheless, one case suggests that some bacteria could have previously possessed this property. Indeed, a cochineal (Planococcus citri, hemiptera) contains two endosymbiotic bacteria, a β-proteobacterium and a ɣ-proteobacterium. It has been shown that the ɣ-proteobacterium actually lives in the cytoplasm of the other bacterium. The mode of penetration of the ɣ-proteobacteria inside β-proteobacteria is not known and the involvement of phagocytosis has not been demonstrated. Currently, only two examples of such symbioses are known, which contrasts with the recurrence of bacterial endosymbiosis in eukaryotic cells.
Back to chapter index