Holomycota
Back to main indexJump to section:
Introduction
The second great evolutionary lineage of Opisthokonta is that of Holomycota, also called Nucletmycea. The most successful group of Holomycota is undoubtedly the Eumycota or “true fungi”. At the base of the phylogenetic tree (Figure 101) are organisms whose mode of nutrition is always phagotrophic, the Nucleariida and the Aphelida, confirming that the Eumycota, whose mode of nutrition is osmotrophy, do indeed derive from phagotrophic protists. This trophic mode also seems to be conserved in Rozella allomycis, a parasite of fungi belonging to Rozellida, the sister group of Eumycota. But the difficulty in cultivating other Rozellida makes their way of life a mystery. Probably encouraged by the lack of information about them, English researchers included them in the Cryptomycota group, without knowing if these organisms are indeed fungi with osmotrophic nutrition! If Rozella’s phagotrophy is typical of the group, it would therefore be better to keep the name of Rozellida to clearly indicate that they are not fungi. Rozellida are related to Microsporidia, a group of intracellular parasites that has regressed to such an extent that for a long time it was considered to be an intermediary between prokaryotes and eukaryotes! Unlike the Eumycota which have been extensively studied, relatively little is known about the other three lineages of Holomycota.
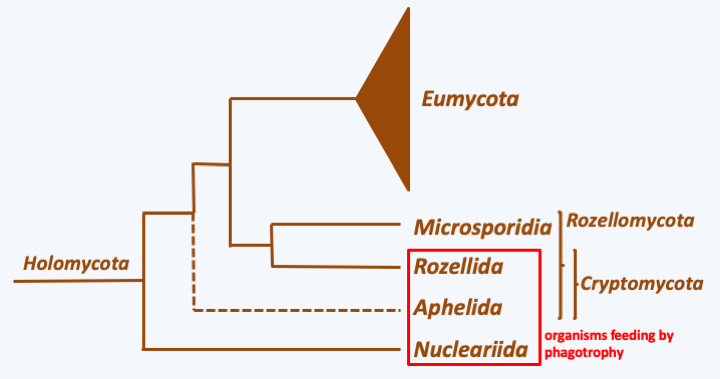
Figure 101.
Phylogenetic tree of Holomycota.Nucleariida
Nucleariida live mainly as amoeba with radiating pseudopodia (Figure 102). These filamentous pseudopods are produced without the support of microtubules, unlike the filopods of other radiating amoebae of the Rhizaria group. Surprisingly, these amoebae have discoid crested mitochondria, like the Discoba. Currently, only a few species are recognized as belonging to this group, but others with similar characters should probably be classified here. Little is known about the group’s biology. Lifestyles and occupied biotopes are not well defined. They were first isolated from a freshwater sample and the species Nucleria pattersoni was obtained from the gills of a fish. Metagenomic data confirms that these organisms are present in many freshwater biotopes. On the other hand, it is possible to cultivate certain species on Petri dishes in monoxenic culture, which should allow their study in more detail. Full life cycles are not well known. Some species seem to live exclusively in unicellular form. They may or may not form cysts. Others, such as Fonticula alba, have developed a rudimentary aggregative multicellularity (Figure 103).
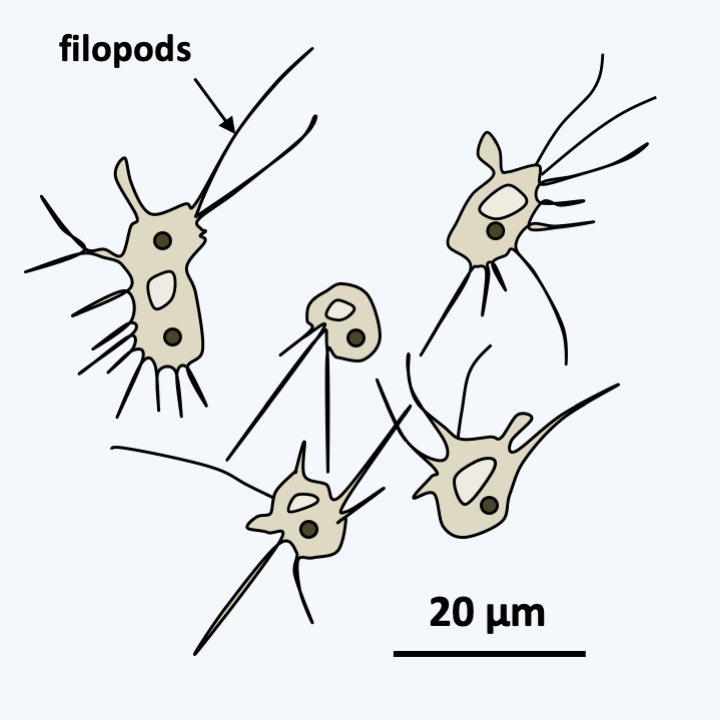
Figure 102.
Nuclearia pattersoni.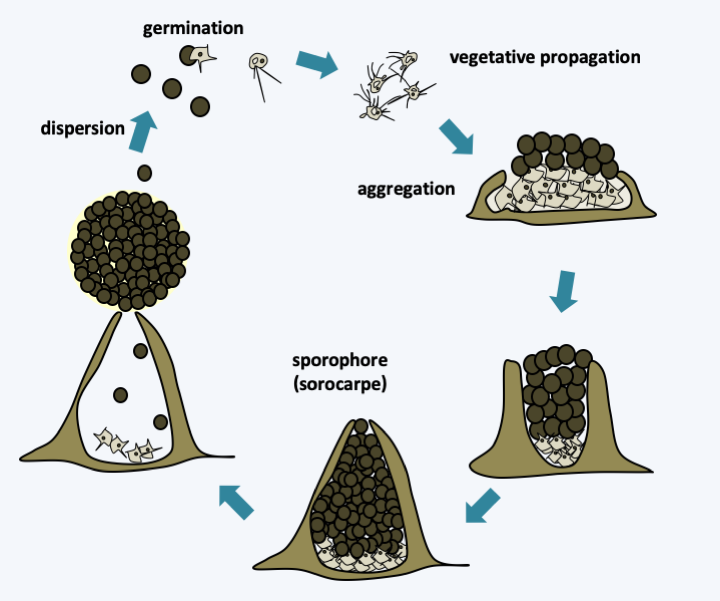
Figure 103.
Life cycle of Fonticula alba. This rare species, being isolated only once, independently established an aggregative multicellularity. It passes its vegetative phase in the form of phagotrophic amoebae differentiating into the typical filopods of Nucleariida. To develop fruiting bodies, free amoeba group together and secrete an extracellular matrix via the Golgi apparatus. Then some of them differentiate into spores which end up being extruded at the top of the sporophore in the form of a sphere giving the sorocarp a characteristic morphology.Aphelida
The few known species of Aphelida are intracellular parasites of unicellular algae whose populations they regulate. The trophic phase is in the form of small phagotrophic amoebae which will give rise to plasmodia (Figure 104). These plasmodia will eventually go through schizogony and produce spores inside the lining of their hosts, which they use for protection. The resulting zoospores can be, depending on the species, either amoeboid or flagellated with the posterior flagellum typical of Opisthokonta. At the base of the flagellum is also a second centriole. There is an immobile pseudo-flagellum in Amoeboaphelidium, the origin of which is obscure. The sequences of some genes show a non-standard genetic code in these microorganisms: UAG and UAA seem to code for glutamine instead of stop.
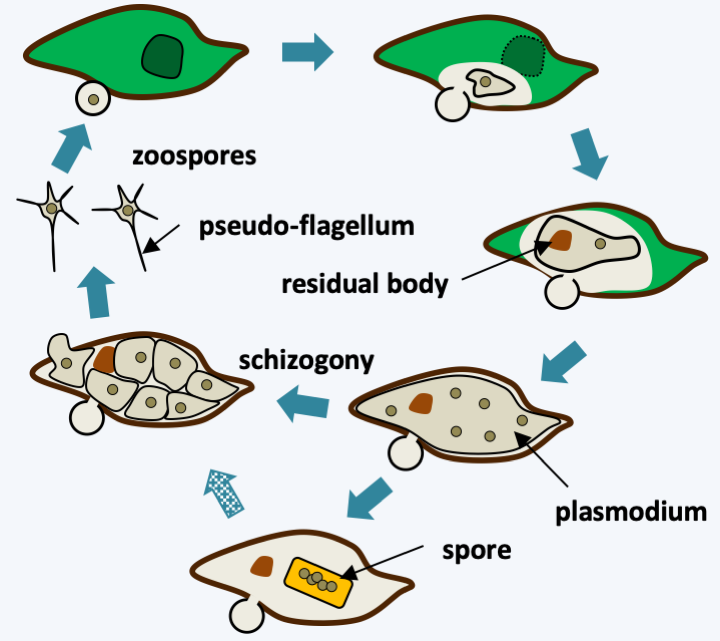
Figure 104.
Life cycle of Amoeboaphelidium protococcarum. Like all aphelids, this species parasitizes unicellular algae. After penetration into its host, the parasite consumes its cellular content and develops a plasmodium, which after schizogony will generate infectious zoospores. In some strains, resistance spores can differentiate. When they germinate, these spores generate zoospores after schizogony. The residual body contains the wastes produced by the parasite.Rozellomycota
The last line which branches before that of the true fungi bears the provisional name of Rozellomycota with two sublineages which are currently well described: the Rozellida and the Microsporidia. It contains a set of species that are very difficult to cultivate for the most part because they are intracellular parasites of other eukaryotes. It is very likely that the phylogeny of this group will shift a lot in the future. Indeed, parasites with similar life cycles and morphologies seem to exist in both subgroups. The reductive evolution often accompanying parasitism means that only molecular phylogenies make it possible to classify them, and even then with difficulty. This is because in one of the two groups, that of Microsporidia, the genome has undergone a reductive (shrinking in size) and accelerated (base substitutions happen at a higher frequency) evolution making it difficult to use its sequence for phylogenetic analysis. Environmental metagenomic data indicate that other, so far undescribed, lineages exist in many biotopes, and that they exhibit significant genetic diversity; maybe as much as the Eumycota!
Rozellida
The Rozellidae have long been known only from about twenty parasitic species of other aquatic fungi, Eumycota or Pseudomycota. More recently, intracellular parasites of amoeba, Paramicrosporidium, showing a strong morphological simplification, and therefore strongly resembling microsporidia, have been described as belonging to this group. We do not currently know the way of life of the species detected by metagenomics and our knowledge of Rozellida therefore derives only from Rozella, of which the best known species is Rozella allomycis (Figure 105). This species is an intracellular parasite of fungi of the genus Allomyces. Its cycle begins with a zoospore comprising a posterior flagellum that attaches to an Allomyces cell. The zoospore retracts its flagellum and becomes encysted. A germ tube forms and enters the interior of the cell. The cell body then passes inside the host cell where it develops at the expense of the cell contents in the form of a naked plasmodium devoid of a chitinous wall. The nutrition of these organisms takes place by phagocytosis of the cytoplasm of the host cell. Then, the plasmodium differentiates into a zoosporangium, the wall of which contains chitin. It is not clear how chitin is synthesized. Initially, it was thought that the host cell provided it, but four genes encoding chitin synthetases have been identified in the genome of Rozella allomycis. This zoosporangium ends up cleaving into zoospores that disperse after rupture of the host cell. The zoosporangium can also differentiate into a resistance sporangium: the life cycle of this species is therefore very simple because sexual reproduction seems absent. The condition as an intracellular parasite suggests that Rozella probably underwent a reductive evolution, making it difficult to extrapolate the knowledge gained on Rozella allomycis to other Rozellida. Hybridization analyzes with various probes on samples taken directly from nature show that other Rozellida are small ovoid cells 2-3 μm in diameter, with a flagellum or lacking a flagellum. The forms without flagellum are interpreted as cysts while the forms with flagellum would be zoospores. The trophic form therefore remains to be discovered. The presence of a chitinous wall typical of Eumycota could not be demonstrated by labeling with a specific lectin, nor the presence of a cellulosic wall. However, chitin has been detected in Paramicrosporidium. In summary, although possessing a life cycle similar to some fungi, especially for some species like Rozella allomycis which produces chitinous walled spores, these organisms probably did not make a complete transition to osmotrophy and therefore cannot be considered true fungi.
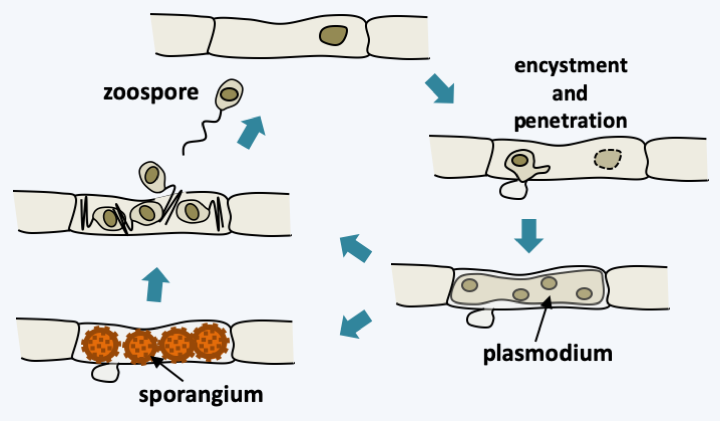
Figure 105.
Life cycle of Rozella allomycis.Microsporidia
The other lineage of Rozellomycota is Microsporidia. Nosema bombycis was the first species described by Pasteur as being responsible for the pebrine disease of the silkworm, Bombyx mori. This disease was wreaking havoc on bombyx breeding centers for silk production. Pasteur showed that these parasites were transmitted to the offspring by the mother during reproduction and developed a treatment which consists of selecting descendants without parasites to initiate new breeding and was thus able to “save French industry Silk”! Since then, more than 1,100 species have been described. It is likely that this figure is very underestimated.
Like Nosema bombycis, other Microsporidia are intracellular parasites of simple structure which infect a large number of eukaryotes, ranging from protists to humans with a preference for invertebrates. They have undergone an important regressive evolution, because they are among the smallest eukaryotic cells and also among the simplest. They do not have peroxisomes nor mitochondria or hydrogenosomes but mitosomes. There are also no cilia or flagella. But they nevertheless have a network of internal membranes (reticulum). They have an underdeveloped Golgi apparatus. This development was accompanied by a decrease in the average size of proteins and a reduction in the complexity of the ribosome, which in size resembles that of prokaryotes. There is also a decrease in the coding capacity of the genome. In Encephalitozoon cuniculi, the genome measures only 2.9 Mb and only encodes around 2,000 genes. Both in size and in coding capacity, this genome is smaller than some bacterial genomes! The genes are very close to each other and the 5′UTR regions are very short, in the order of three to ten nucleotides. Duplicate genes are rare. In this species, there are no genes involved in the citric acid cycle and the respiratory chain, and few genes are involved in the biosynthesis of amino acids. On the other hand, there are many transporters, in particular translocases, making it possible to import nutrients including ATP from the cytoplasm of the host cell. In contrast to Encephalitozoon cuniculi, the genome of Mitosporidium daphniae measures 5.7 Mb and encodes 3,300 genes. This species experienced less regression because during the sequencing project, a mitochondrial genome could be identified. The genes, present either on the mitochondrial genome or on the nuclear genome, still seem to allow partial production of ATP without knowing the final electron acceptor. Correspondingly, the genome does not appear to encode the ATP/ADP translocases that allow other Microsporidia to take ATP from their host.
The Microsporidia life cycle can be simple or complex depending on the species. In particular, there is in some species a sexuality whose modalities are not completely elucidated, as evidenced by the presence mating type genes in their genome. Likewise, some Microsporidia need several successive hosts to complete their entire cycle. In Enterocytozoon bieneusi, sexuality is unknown and the life cycle is simple (Figure 106). Infectious spores are ingested. They come into contact with the host cell. They extrude an organelle, called the polar tube, which passes through the cell membrane, allowing entry into the host’s cytoplasm. It all happens in 2 seconds! The spore first undergoes morphological changes, notably with swelling of its vacuole; this is due to the increased osmotic pressure in the spore. This increase is because the spore, which is impermeable to water, is very concentrated in solutes. The activated spore hydrolyzes certain polymers, which leads to a peak in osmotic pressure which will then allow water to enter. Then a rupture of the anchor disc occurs. The polar tube is then violently expelled, allowing it to cross the plasma membrane of the host cell. The sporoplasm, that is, the cellular content of the spore, passes through the polar tube to the host cell; note the smallness of the polar tube and the deformation that the sporoplasm must therefore undergo! The mechanics of the passage of the sporoplasm is again probably done by osmotic pressure with an increase in size of a vacuole which will remain in the spore. In the host cell, the sporoplasm undergoes massive multiplication by binary fission. In some species, therefore, this process can give rise to cells with multiple plasmodium-like nuclei or to more complex arrangements with multiple cells with multiple nuclei. This multiplication can take place either in direct contact with the cytoplasm of the host cell or in a parasitophorous vacuole. This proliferation will give rise to what are called meronts. The latter will undergo a stage of division and maturation: sporogony. These are at this stage called sporonts. This phase allows the sporonts to acquire all the structures of the spore such as the spore capsule or the polar tube… This is when meiosis could occur in some species. The sporogony phase can also lead to xenoma at the end of the infectious phase. Xenoma is a complex in which intracellular microsporidia take metabolic control of the host cell to increase spore production (Figure 107). For example, it is not uncommon to find arthropods in which almost the entire organism has been replaced by an enlarged mass of Microsporidia spores. The spores are surrounded by a wall containing proteins, polysaccharides and chitin. They are very resistant to variations in temperature, pH, etc., and therefore effectively disperse these parasites.
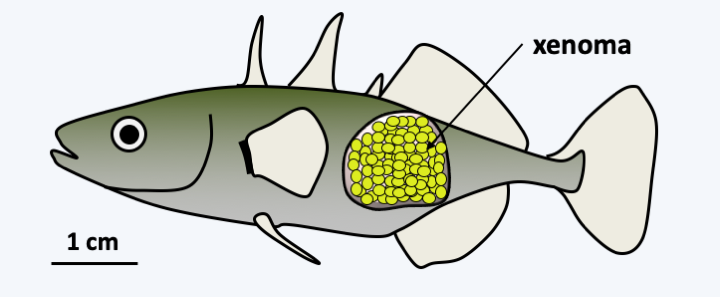
Figure 107.
Xenoma caused by Glugea anomala after infection of a stickleback (Gasterosteus aculeatus).It should be noted that some Microsporidia have very wide host spectra and that the same species can infect both vertebrates and invertebrates. They pose many problems in insect farms, including Nosema apis which parasitizes bees and of course Nosema bombycis which attacks silkworms. Some insect parasitic species could be used as biopesticides, in particular Nosema locustae against locusts or Brachiola algerae against mosquitoes. Other important targets for farming include crustaceans, molluscs and fish, birds… Some species are opportunistic pathogens that frequently infect immunocompromised people and are responsible for “microsporidiosis”. Table 8 summarizes the main human infections. It is estimated that 8% of the European population has been infected once in their lifetime with Microsporidia. In most cases, the infection remains asymptomatic.
Back to chapter index