Difficulty of the notion of species in eukaryotic protists
Back to main indexThe nomenclature of organisms is based on the principles proposed by Carl Von Linné (1707-1778). Each individual must be able to be identified by its genus and species name. Indeed, Linnaeus proposed a classification system which groups together organisms which resemble each other in increasingly large sets (Table 6). Note that the seven levels defined by Linnaeus (from the broadest to the most restricted: kingdom, phylum, class, order, family, genus and species) are now very insufficient to correctly classify all species, so a very large number of intermediate levels: super- and sub-phyla, infra-sub- and super-classes, etc. are now also in use. Nevertheless, the basic unit of this system, or of other more modern related systems such as cladistics, remains the species, which is denoted by two adjoining substantives: “Genus species”.
Classically, a species is defined as a set of inter-fertile individuals, that is to say capable of interbreeding and giving birth to fertile offspring. While this definition applies relatively well to animals and plants, it does not apply to eukaryotic microbes. In fact, in many cases of “lower” eukaryotes, sexuality is not known, or is even non-existent, making it difficult to determine membership of the same species. In this case, molecular analyzes can reveal genetic associations or haplotypes. The analysis of these haplotypes and the way in which they recombine in populations makes it possible to define different species as isolates that do not recombine at the genetic level (Figure 66). Take note however, that these analyzes will not take into account horizontal transfers, by which even very phylogenetically distant species can “exchange” genes; often the transfer is unidirectional. Indeed, horizontal transfers are generally rare events in long-term evolution. On the other hand, haplotype analyses will take into account the exchange of genetic material that takes place not only sexually but also “parasexually”. Indeed, the reciprocal exchange of genetic information between individuals of the same species can take place in non-sexual forms in many Eumycota fungi or in protozoa such as Dictyostelida (Figure 67).
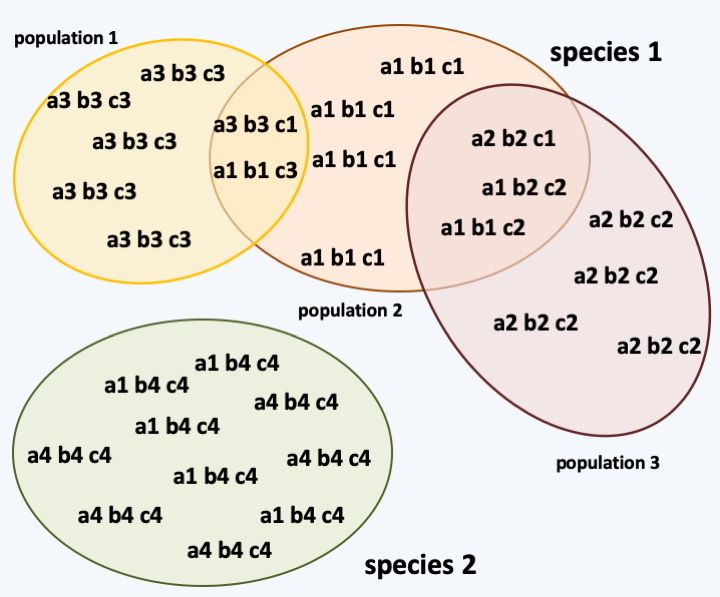
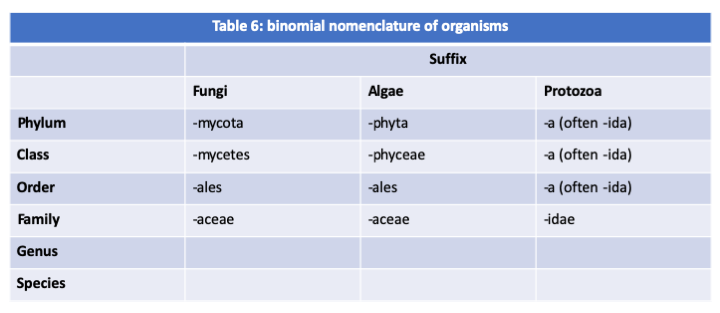
Figure 66.
Analysis of haplotypes for the definition of species. Species # 1 has three populations that exchange genetic information, as evidenced by the presence of numerous recombinants. Species # 2 has only one population that does not exchange information with those of Species # 1, although they share the a1 marker. The discovery of individuals with recombinant haplotypes, such as 'a4 b1 c1' or 'a3 b3 c4', would allow the conclusion that species # 2 is just another population of species # 1. The polymorphic markers used for these studies are diverse: nucleotide differences in coding sequences, microsatellites, indel, etc.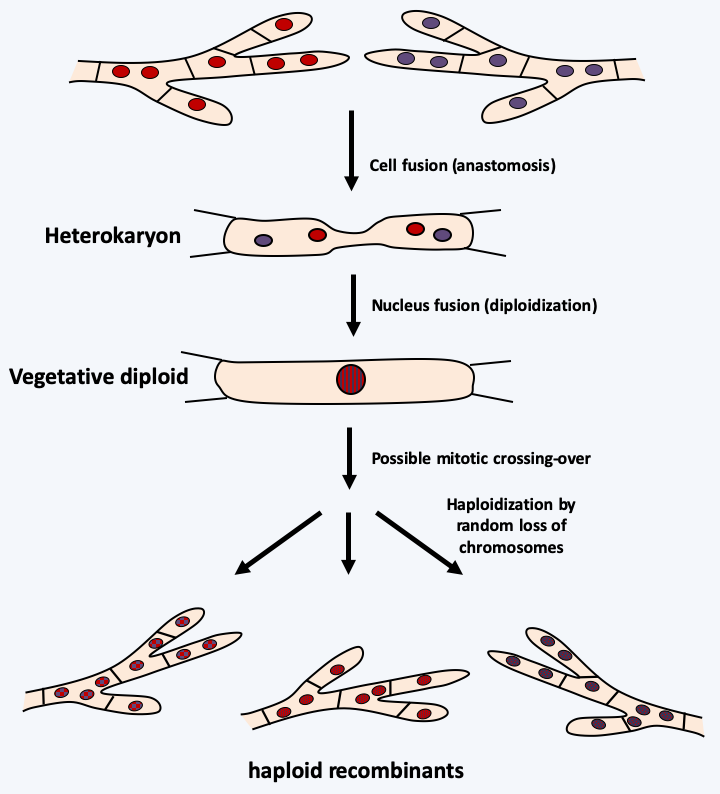
Figure 67.
The para-sexual cycle. This mechanism allows an alternation of haploid and diploid generations without sexuality. It begins with an anastomosis, that is, the plasmogamy of two haploid cells without gamete formation. The resulting heterokaryon can undergo karyogamy (fusion of nuclei) to give diploid nuclei. These are very unstable and quickly give back after random loss of chromosomes from the haploid nuclei. The recombinants thus obtained will present a random mosaic of the chromosomes of the two “parents”. DNA damage repair events also sometimes lead to mitotic crossovers in the diploid nucleus, which will add to the genetic diversity of the recombinants. We do not know the frequency of these events in nature, but this mechanism is easily demonstrated in the laboratory.As species (at least their populations) are subject to Darwinian evolution, it is not uncommon to encounter populations that have genetically diverged. This can result in a decrease in fertility, or even sterility, between individuals from distant populations, but not between individuals from close populations, which is visualized by a low rate of recombination of haplotypes (see Figure 66 where populations 1 and 3 no longer exchange information, except via population 2). We then speak of a complex of species to designate all of these more or less inter-fertile populations. This type of population structuring appears to be the rule rather than the exception in many eukaryotic protists. Likewise, there are many proven cases, in Eumycota for example, where a new species actually results from the hybridization of two species. For example, the ascomycete fungi Neurospora crassa and Neurospora intermedia are considered to be distinct species on different morphological and biological criteria. However, it is possible to cross them with each other in the laboratory and to obtain offspring. Many crosses are required and the spore yield is low. The resulting strains are fertile and have one half of the genome from Neurospora crassa and the other half from the genome of Neurospora intermedia. These hybrids can cross quite effectively with the two starting species. Hybridizations are not laboratory artifacts because many species derived from hybrids have been demonstrated in Saccharomycotina yeasts or fungi of the Epichloë/Neotyphodium group, among others. In some cases, this can have an impact on our societies. For example, the virulent ascomycete fungus Ophiostoma novo-ulmi which currently destroys elms in Europe and North America originated from a hybrid of Ophiostoma ulmi which had previously ravaged elms and from which it took the mating type MAT-1 and genes regulating cell fusions, and a less virulent strain of Ophiostoma novo-ulmi. In this case, the infertile hybrid that served as a bridge between the two species was found in the area of confrontation between the Ophiostoma ulmi and Ophiostoma novo-ulmi species. The removal of barriers that limit hybridization suggests that these are not very difficult to cross. In fact, the deletion of the sad-1 gene which is involved in a mechanism of gene silencing acting in meiosis, related to RNA interference, allows the cross between Neurospora crassa and Neurospora intermedia with fertility levels similar to those of intra-species crosses. These two species do not cross because they differ in some chromosomal rearrangements which are the target of meiotic inactivation. Nevertheless, even if the species complexes and hybrids make the notion of species more vague, the criterion of isolation of genetic recombination takes more and more prominence for the definition of species in eukaryotic microorganisms, in particular if true sexuality, that is to say the succession of meiosis and fertilization, is not observed.
From a practical point of view, if the criteria for reproductive isolation are lacking to easily define a species, and the analysis of the genetic structure of populations of microorganisms requiring a prohibitive lot of work and money, then we define morphospecies as individuals having the same morphology. Often these morphospecies group together several species in the sense of a recombination group. For example, in Paramecium caudatum, which is a sexual morphospecies of Ciliophora, the analysis of crosses shows that it is possible to divide this morphospecies into at least 16 distinct species. Likewise, the amoeba Entamoeba histolytica has long mystified biologists, because some strains appear to be pathogenic to humans and others not. This species was in fact a complex of two species: Entamoeba histolytica and Entamoeba dispar. They are morphologically identical but the first one is pathogenic and the second one is not. Numerous polymorphisms between the two species have been detected by the sequence of their genomes, confirming that they are indeed two distinct species. In fact, the mean difference between the orthologous regions of Entamoeba histolytica and Entamoeba dispar is 38% for the synonymous sites, while the mean nucleotide divergence between Entamoeba histolytica strains is 0.05%. For comparison, the average nucleotide difference between humans and chimpanzees is in the order of 1 to 2%. The availability of genome sequences now provides a “final” criterion for differentiating between species. Indeed, it is accepted that if organisms have a nucleotide polymorphism of their genomic sequence less than 1%, it is probably the same species, if it is greater than 5%, they are two distinct species. If this percentage is between 1 and 5%, further analyzes will be necessary to decide!
Back to chapter index