Plastids
Back to main indexJump to section:
Introduction
Like the mitochondria, the plastids of photosynthetic eukaryotes, algae and plants, most likely derive from an endosymbiosis: that of a cyanobacterium with a eukaryotic cell already possessing mitochondria, to give what is called an alga. However, the history of plastids is much more complex than that of mitochondria, as there appear to have been at least two independent initial endosymbioses. In addition, one of the two was followed by additional symbioses, where eukaryotic algae will in turn act as endosymbiosis partners to give rise to new lineages of algae. We talk about the event of initial cyanobacterial endosymbiosis called primary endosymbiosis. The other endosymbioses are termed secondary, even in certain cases tertiary or quaternary! The main argument supporting these assertions is similar to that of mitochondrial endosymbiosis: the plastids from primary endosymbiosis resemble photosynthetic bacteria having degenerated, and those from secondary endosymbiosis to eukaryotic algae having degenerated.
Primary endosymbioses
Plastids from primary endosymbioses have two membranes and have a genome which codes, among other things, for subunits of photosynthetic complexes. Among the different lineages resulting from primary endosymbiosis, one, that of Glaucophyta (Figure 34) even contains plastids, called cyanelles in these organisms, which have conserved peptidoglycan between their two membranes and the photosynthetic pigments typical of cyanobacteria! Comparisons of the genomes of plastids from primary endosymbiosis with those of bacteria (Figure 35) show that they are related with the cyanobacterial genomes of group A and that all have a common origin, except the plastid present in Paulinellida “Paulinella chromatophora” which is related to cyanobacteria of the C group. The origin of this plastid, called the chromatophore, is much more recent than that of the others and its study provides a better understanding of the first steps in the establishment of this type of symbiosis (Box 6). There are therefore two origins to plastids, one which gave almost all of the current plastids and which would have occurred between one and two billion years ago and the other much more recent dating from around sixty million years ago. Certain group A cyanobacteria, to which the usual plastids are related, are able to fix nitrogen, which no current eukaryote seems to be able to do (Table 1), except perhaps Paulinella chromatophora, because the genes encoding the nitrogen fixation are present in the chromatophore genome. The eukaryotic host of the first endosymbiosis does not seem to have left any descendants that would not have made the primary endosymbiosis because all Archaeplastida, the monophyletic line which includes the organisms resulting from the primary endosymbiosis, possess or at least have possessed a plastid. Analysis of the genomes of Archaeplastida shows that they contain an unusually high number of genes resembling those of Chlamydiae, obligate intracellular parasitic eubacteria of eukaryotic cells. It is therefore possible that Chlamydiae were present in the host before the arrival of the cyanobacterium and that they may have helped establish the symbiosis.
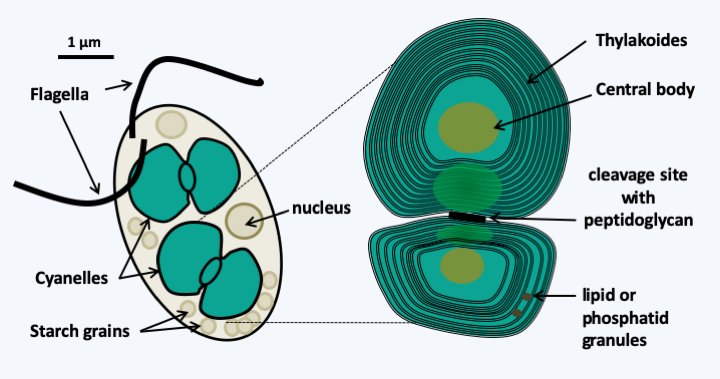
Figure 34.
Cyanophora paradoxa and its cyanelles. Cyanophora paradoxa is a motile glaucophyte alga living in freshwater. It has a beautiful blue-green color due to the presence of cyanelles, a shade found in cyanobacteria and which is due to their photosynthetic pigments. The internal structure of cyanelles closely resembles that of cyanobacteria, with the presence of peptidoglycan between the two membranes clearly visible at the level of the plastid cleavage site. The central body can be likened to a carboxisome which is the site of CO2 fixation in prokaryotes; the equivalent in other plastids is called pyrenoids. The initial descriptions of these cyanelles defined them as endosymbotic cyanobacteria. The inability to grow them in axenic cultures and then the genome sequences contained in the cyanelles clearly show that they are plastids.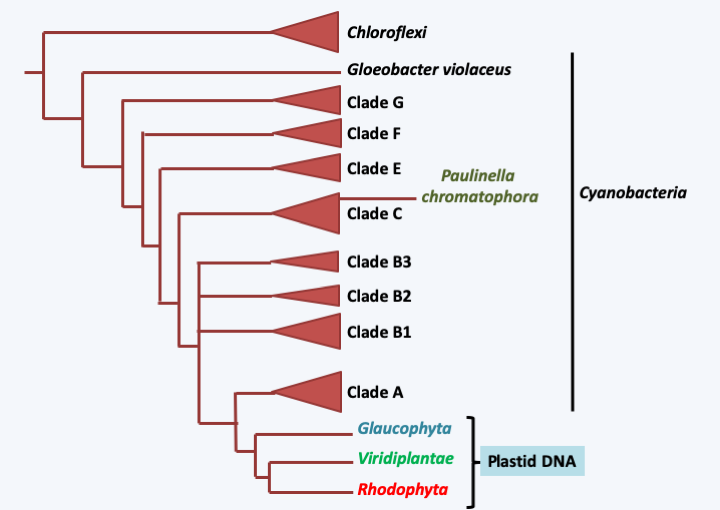
Figure 35.
Phylogenetic tree of plastid genomes from primary endosymbiosis and cyanobacteria. Plastids have a common origin and are related to cyanobacteria of groups A and B1, except the chromatophores of Paulinella.Box 6. The chromatophore of Paulinella
Amoeba belonging to the Paulinella chromatophora species complex (there are at least two morphologically and genetically distinct lineages of this 'species') have a 'plastid' called a chromatophore, whose origin is different from all other plastids known today. Like other non-photosynthetic and phagotrophic members of the genus Paulinella, these algae are Rhizaria amoebae that live in freshwater and are protected by siliceous scales. The sequence of the chromatophore genome shows that it strongly resembles that of cyanobacteria. However, it underwent a reductive evolution as it measures only one megabase and encodes only 867 proteins, 42 tRNAs and ribosomal RNAs. This corresponds to a coding capacity three to four times less than that of a typical cyanobacterial genome. Based on a typical loss rate for an intracellular symbiont, the age of the chromatophore has been estimated to be 60 million years; the estimated age for primary endosymbiosis that gave rise to the other plastids is 1.5 billion years. Among other things, this genome has lost genes involved in photosynthesis, the biosynthesis of amino acids and cofactors. Some of these genes are relocated to the nucleus, where they have acquired characteristics specific to nuclear genes; in particular, some have introns typical of the nucleus. The proteins they encode are imported into the chromatophore, showing that it really is an organelle and not an endosymbiotic bacterium. The import system appears to be different from that of classical plastids because the proteins pass through the reticulum and the Golgi apparatus; a route also chosen for trafficking to certain plastids from secondary endosymbioses. Other symbioses between eukaryotic protists and cyanobacteria exist. Studies are underway to see if, like the chromatophore, the cyanobacteria have a different origin than that of the plastid and to what extent they are integrated into the cellular physiology of the host.
Plastids from primary endosymbiosis are surrounded by a double membrane and as in the case of mitochondria, the origin of the double membrane is not clearly understood. Does it correspond to the double membrane of cyanobacteria or to the membrane of the endocytosis vesicle and the internal membrane of cyanobacteria? The question remains open, although the structure of the cyanelle in Glaucophyta suggests that the first solution is the correct one. The selection pressure that favored the first primary endosymbiosis is very probably linked to the photosynthetic capacity of the initial cyanobacterium. Indeed, the fact that no Archaeplastida possesses nitrogenase indicates that the possibility of fixing atmospheric nitrogen is not involved in the establishment of the symbiosis. Initially, cyanobacteria were to serve as prey. But, like all photosynthetic organisms, cyanobacteria secrete sugars into the environment around them. In fact, photosynthesis leads to the formation of a gradient of sugars, free or in complex with other molecules, such as adenosine diphosphoglucose, from the inside to the outside of the cell. The diffusion of sugars to low concentrations to the outside of the cell is an inevitable phenomenon. This is the same phenomenon that allows, for example, the growth of many microorganisms around the roots of plants, a place called the rhizosphere. The persistence of cyanobacteria in the cytoplasm of the predator therefore results in immediate benefit. The sugars thus captured could have been polymerized in the form of glycogen, reinforcing the gradient. The presence of Chlamydiae would have facilitated the establishment of the symbiosis by providing enzymes to polymerize sugars, transporters facilitating the diffusion and then the export of sugars outside the cyanobacteria and the escape of the digestion vesicle, where the cyanobacteria could have coexisted with the Chlamydiae, towards the cytosol. The eukaryotic host would also have contributed to the metabolic integration of the plastids by providing transporters and enzymes allowing the storage of sugars, in particular in the Viridiplantae, in which starch synthesis has been relocated to the plastid. In the Glaucophyta and Rhodophyta starch production remained in the cytosol. The current photosynthetic metabolism, in particular the pathways that serve as storage for starch, therefore result from a complex mixture of enzymes and transporters from all three partners. In addition to photosynthesis, plastids perform a large number of metabolic functions including the biosynthesis of amino acids and nitrogenous bases, purines and pyrimidines. Once in place, the first primary symbiosis followed the same evolutionary paths as that of the mitochondria: loss of genes to the nucleus and relocation of plastid proteins via an import system, the origin of which also seems to be bipartite: the heart would come from the cyanobacteria and the annex proteins from the host. About 18% of Arabidopsis thaliana genes are thought to be of cyanobacterial origin. But since the endosymbiosis is more recent than that giving rise to mitochondria, the transfer is less extensive. For example, 127 genes are present in the genome of Nephroselmis olivacea. The sizes of the plastid genomes are therefore larger and the structures more complex than those of the mitochondrial genomes. Gene transfers to the nucleus, which can also be easily demonstrated in the laboratory, have been accompanied by changes in size (in part via the presence/absence of group I and II introns) and coding capacity of plastid genomes (Figure 36). While in many Archaeplastida, the plastids provide photosynthetic function, this has been lost in non-photosynthetic algae and plants, most of which live as parasites. This is the case, for example, with Prototheca, which infect mammals including humans, or Helicosporidium, which parasitizes the intestines of insects. In these two genera, the plastid genome sequence shows a slight reduction in coding capacities because most of the genes encoding the subunits of photosynthetic complexes are still present (Figure 36), suggesting a recent adaptation to parasitic life. In the case of Polytomella, “green algae” without photosynthesis related to the genus Chlamydomonas, the plastid genome appears to have completely disappeared, although the organelle is still present (Figure 37). The genes involved in the expression of plastid genes, such as those encoding ribosome proteins functioning in the organelle, have also disappeared from the nuclear genome! This reduction is reminiscent of that of the mitochondria into a mitosome. In addition to the modification of the genome and the physiology that accompanies it, the expression of genes could evolve, as in the case of mitochondria, towards non-canonical modes of operation: trans-splicing and editing of mRNAs are common. However, these derived modes of expression are less prominent than in mitochondria. For example, there are no real changes in the genetic code. As in cyanobacteria and other prokaryotes, translation initiation can occur on codons other than AUG, but no codons have actually been reassigned.
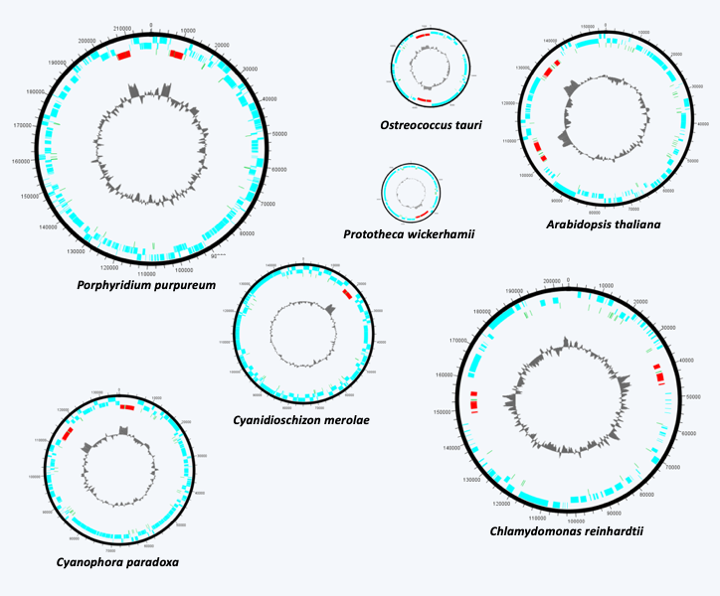
Figure 36.
Structure of some plastid genomes from primary endosymbiosis. Plastid DNAs from primary endosymbiosis are all believed to be circular. The number of genes is in the order of a hundred. They encode the plastid translation system and various subunits of photosynthetic complexes. Long intergenic regions and group I and II introns are often present.Like mitochondria in the case of eukaryotes, plastids from major primary endosymbiosis can serve as a phylogenetic marker to trace the evolutionary history of Archaeplastida. Previously, photosynthetic pigments (chlorophylls, carotenoids and phycobilins) were used as a basis for classification, but there are many exceptions which generally make these pigments poor phylogenetic markers. Analyzes of gene or genome sequences show that Archaeplastida evolved as three distinct lineages. The first to have diverged is that of Glaucophyta (Figure 35); it retained the same photosynthetic pigments as the cyanobacteria, just like the Rhodophyta (Table 4). However, the ancillary pigments (including phycoerythrin) present in Rhodophyta allow the capture of the most energetic visible photons of green and blue light, and therefore penetrate deepest in water, while those of Viridiplantae do not trap these photons. They trap the red and blue-violet lights. The Rhodophyta will therefore be able to grow deeper, in particular along the coasts, where they mainly occupy the lower level located several meters deep. The last line, that of Viridiplantae, abandoned chlorophyll c and phycobilins to adopt chlorophyll b (Table 4).
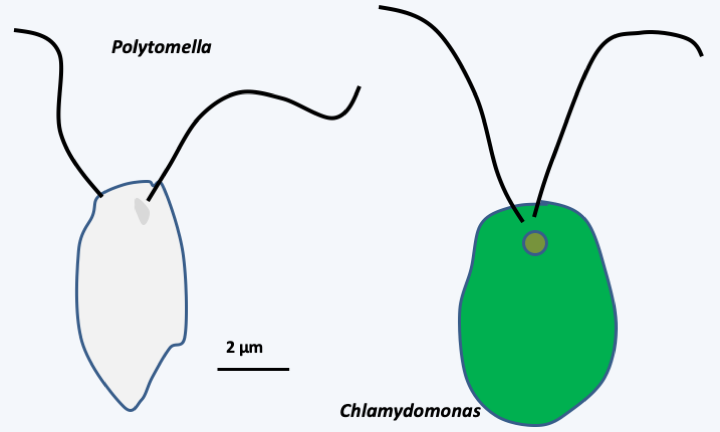
Figure 37.
Polytomella and Chlamydomonas. These two algae are related but one is photosynthetic and the other is not. Although without photosynthetic pigment, Polytomella has a vestige of plastid.Secondary, tertiary endosymbioses, etc.
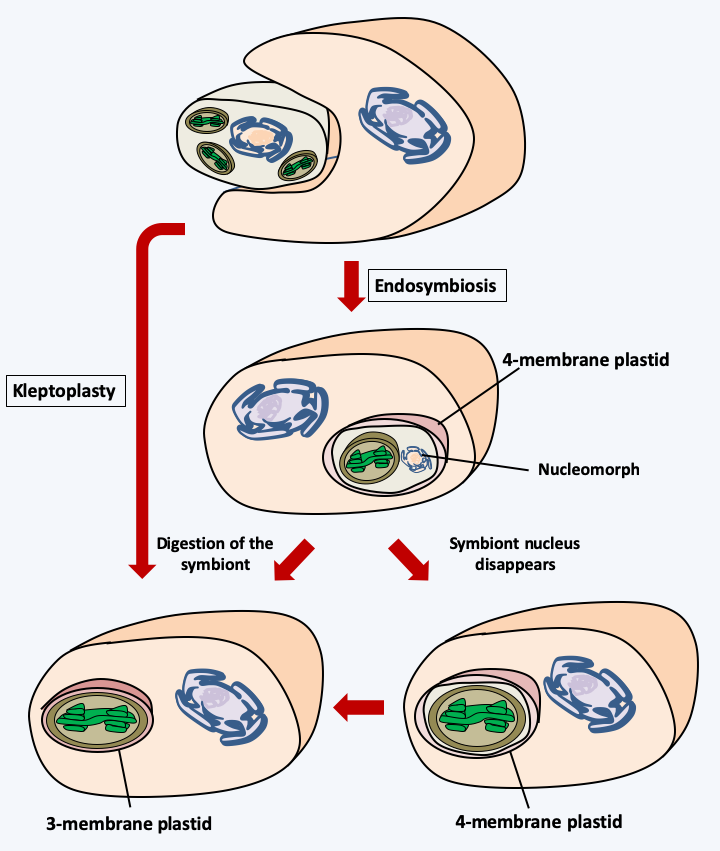
Figure 38.
Mechanisms of appearance of plastids with 3 or 4 membranes. Kleptoplasty (Box 7) involves the incomplete digestion of an Archaeplastida alga, green or red depending on the case, and the retention of plastids within the digestion vesicle. The durability of the plastids therefore subsequently gives an organization with three membranes. Endosymbiosis involves a more progressive association of the two partners. This is accompanied by a regression of the nucleus of the symbiote which becomes a nucleomorph (Box 8). The loss of the nucleomorph results in a plastid surrounded by four membranes. If the plastid escapes from the endocytic vesicle or if the plasma membrane of the symbiont disappears, like kleptoplasty, a three-membrane plastid is obtained. Other more complex mechanisms have been proposed to explain the organization of the three- and four-membrane plastids.The complexity of the history of plastids quickly became apparent to biologists. In fact, the observation of plastids from various groups of algae by electron microscopy has shown that, in addition to plastids surrounded by two membranes, there are plastids surrounded by three or four membranes (Figure 38). The simplest explanation for this situation is that these plastids are derived either from plastid theft events or kleptoplasty (Box 7), or from secondary endosymbiosis involving eukaryotic algae as the photosynthetic endobiont. In Cryptophyta and Chlorarachniophyta, the presence of a degenerate or nucleomorphic nucleus (Box 8) associated with reticulum within the plastid proves that in these two groups the plastid is the result of secondary endosymbiosis. The number of these secondary endosymbiosis events is still debated. Indeed, the initial photosynthetic partner could have been either an alga belonging to either of the Rhodophyta or Viridiplantae, while the hosts have very diverse phylogenetic origins. In the case of endosymbioses involving Viridiplantae, the scientific community agrees on a number of two independent secondary symbiosis events (Figure 39andFigure 40). On the other hand, in the case where the symbiosis involved a Rhodophyta, disagreement reigns with scenarios involving one to several independent initial events, or even a succession of events (Figure 40). There are many reasons for such diverse scenarios to explain the genesis of large groups of algae. The presence of species with or without plastids within the same group could be linked to the loss of plastids rather than to their acquisition. This phenomenon of plastid loss could be widespread and explain the discrepancies observed between the presence/absence of plastids and the phylogenies obtained with nuclear or mitochondrial genes. For example, in the genomes of Oomycota and Blastocytis hominis, a parasite related to Oomycota and Ochrophyta algae, there are genes of cyanobacterial origin, confirming that these organisms probably derive from algae, although they are not found with any relic of a plastid. Two independent losses are nevertheless necessary to explain the phylogeny (Figure 40), which leads some researchers to support the hypothesis that genes of cyanobacterial origin rather result from horizontal transfers from cyanobacteria which would have served as prey for the ancestors of the Oomycota and Blastocystis. In Apicomplexa, the plastid has regressed and no longer performs its photosynthetic function. In these organisms it is called “apicoplast”. In contrast, in Cryptosporidium hominis, an Apicomplexa which rapidly diverged from other parasitic Apicomplexa, the apicoplast has completely disappeared. But genes encoding proteins that function in the apicoplast in other Apicomplexa are still present in the nuclear genome, suggesting that the ancestors of this species did indeed possess a plastid. In contrast, in Ciliophora which are also related to Apicomplexa, genes from cyanobacteria are absent, indicating that they probably never had plastids, thus rejecting the hypothesis of a single secondary endosymbiosis with a Rhodophyta. Another point supporting the multiple scenarios hypothesis is that it is not because two closely related lineages have plastids that they have a common origin. For example, the different plastid structures of Apicomplexa with four membranes and Peridinal Dinoflagellata with three membranes suggest that contrary to what is noted inFigure 40, the plastids could result from independent events in the two groups. The alternative hypothesis is that, as noted inFigure 40, the origin is quite common, but in Dinoflagellata the plastid may have emerged from the endocytic vesicle during evolution, but not in Apicomplexa. Current data, including recent phylogenies on a large number of genes, rather favor the hypothesis of multiple secondary endosymbiosis events associated with serial kleptoplasty and recurrent losses!
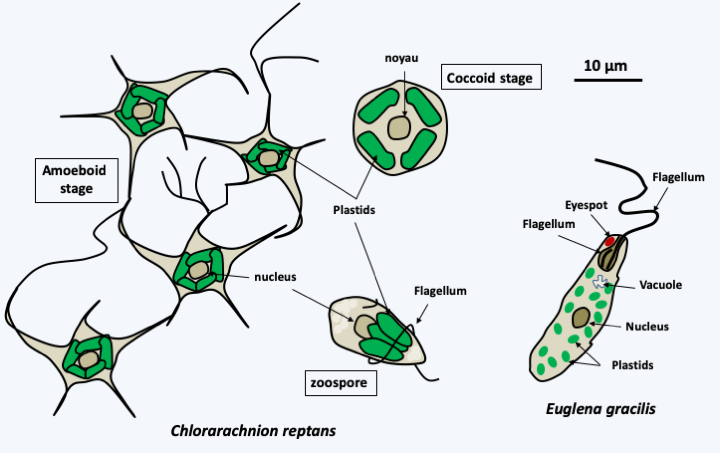
Figure 39.
Chlorarachniophyte algae and photosynthetic euglenids are derived from secondary endosymbiosis events independent of Viridiplantae algae. Chlorarachnion reptans is a typical chlorarachniophyte whose plastid is surrounded by four membranes and always contains a nucleomorph (see Box 7). It can take three distinct forms depending on its stage of development. The amoeboid form can still feed by phagotrophy. Sequence analyses of the plastid genome and photosynthetic pigments indicate that it is derived from a Viridiplantae related to the genera Chlamydomonas and Chlorella of the 'TUC' clade. Photosynthetic euglenids, such as Euglena gracilis, are related to non-photosynthetic euglenids and feed by phagotrophy. These algae have a simple cycle with no known sexual reproduction and their plastid is surrounded by three membranes. They can lose it and feed by pinocytosis and osmotrophy; they often live in environments loaded with organic matter. Indeed, losses in photosynthetic capacity associated with the presence of degenerated plastids have occurred several times independently during the evolution of euglenids. Phylogenies based on the genome sequence of the plastid show that it is derived from a Viridiplantae related to Prasinophyta.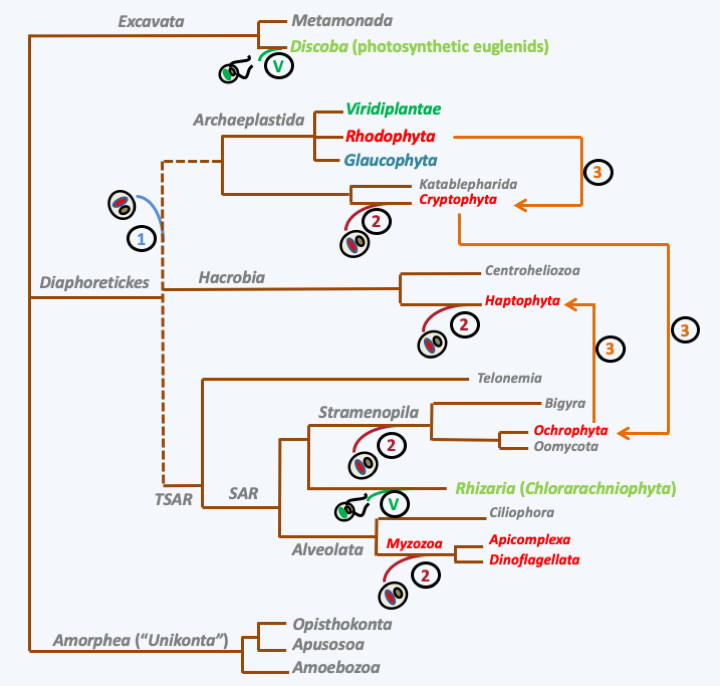
Figure 40.
Scenarios of secondary endosymbiosis. Endosymbiosis with Viridiplantae (V) algae occurred twice independently during evolution. The number of endosymbioses with Rhodophyta is still a matter of debate. In Scenario 1, a single secondary endosymbiosis gave rise to all the groups of algae carrying such plastids, which are all in the great diaphoretickes super-phylum. According to this scenario, the groups (in gray) without plastid lost it, which necessitates recurrent and numerous losses. In Scenario 2, four secondary endosymbiosis events have occurred. In this scenario, there is also a need for plastid loss, for example in Oomycota and Bygira. In Scenario 3, successive endosymbiosis occurred: secondary endosymbiosis of a Rhodophyta gave rise to Cryptophyta, which was engulfed in a tertiary endosymbiosis event to give rise to Ochrophyta algae. An Ochrophyta in turn made a quaternary endosymbiosis to give the algae Haptophyta. In this scenario, it is possible that it is serial kleptoplasty (see Box 6) rather than true endosymbiosis.Box 7. Kleptoplasty
Kleptoplasty is the theft of plastids by another organism. After ingestion of an algae, it is incompletely digested and its plastids often persist within vesicles in the predator. This mechanism is therefore different from an endosymbiosis where the algae remains intact inside its host. The best-known example is that of the sea slugs of the genus Elysia. Depending on the species, these slugs keep the plastids of the algae that they eat from a few days to over a month. Elysia chlorotica is the most studied.
Like all Elysia, Elysia chlorotica is born without plastids and in order to acquire them, the larvae must consume Stramenopila algae of the Vaucheria genus for a few days. In this symbiosis, the plastids of Vaucheria are not digested and are stored in cells which surround the digestive system, which is very folded. Plastids provide part of the carbonaceous nutrition during the 10 months of the life of this organism. Note that this gastropod is very flat, like... a leaf! However, while these slugs resist deficiency for a long time, it appears that plastids are not sufficient to provide complete food independence, and slugs continue to feed throughout their lives. The plastids do not divide, which is why they are not passed on to the offspring. The presence of genes from Vaucheria in the Elysia genome remains under debate, as conflicting results have been obtained. It appears that algal DNA can persist in a non-integrated form in the animal's genome and therefore would not be transmissible to offspring. This could explain why plastids persist for so long while remaining functional.
Elysia are not the only organisms capable of kleptoplasty, because it is found in Foraminifera and especially Dinoflagellata. The process can be observed in Dinoflagellata strains related to the genera Karenia and Karlodinium, tentatively referred to as RS-24 or W5-1. Analyses of the plastid genomes of RS-24 and W5-1 show that these genomes have a different origin from those of Karenia and Karlodinium, although all are derived from Haptophyta. Indeed, they result from kleptoplasty of the two plastids of Phaeocystis antarctica, whereas in Karenia and Karlodinium the plastids are the result of an older symbiosis. Cultivation of RS-24 and W5-1 is only possible in the presence of Phaeocystis antarctica. Division of RS-24 and W5-1 cells, but not their survival, requires light even if prey is present in the dark. Without the addition of Phaeocystis antarctica, RS-24 and W5-1 gradually lose their plastids, the number of which stabilizes at three if prey are still present in low numbers. It goes back to 20 as soon as prey is added in large numbers.
The most extraordinary example, however, is that involving Cryptophyta of the genera Geminigira/Teleaulax, a Ciliophora (Myrionecta rubra) and Dinoflagellata of the genus Dinophysis including Dinophysis acuminata and Dinophysis fortii. Geminigira cryophila and Teleaulax amphioxeia serve as prey for Myrionecta rubra. The latter species retains not only the plastids of its prey but also their nuclei, which remain transcriptionally active and provide mRNAs to ensure the maintenance of the plastid in the ciliate over a very long period. This nuclear theft is called 'karyokleptia'. Myrionecta rubra in turn serves as prey for Dinophysis acuminata or Dinophysis fortii. These two Dinophysis also contain a plastid whose structure is different from that of Geminigira cryophila but whose DNA is identical. It has lost the outer membrane and the nucleomorph, its pyrenoids are distinct. Dinophysis acuminata can only be grown in the presence of Myrionecta rubra as prey, but not with Geminigira cryophila! It is therefore possible that the Dinophysis are stealing the plastid of Myrionecta rubra, itself stolen from Geminigira cryophila. An alternative hypothesis would be that it takes in Myrionecta rubra factors necessary for the maintenance of the plastid, such as transcripts of the Geminigira cryophila nucleus. The plastid would have been acquired previously during its evolutionary history by kleptoplasty either from Geminigira cryophila or from Myrionecta rubra!
Confusion over the evolution of plastids is even more prevalent if Dinoflagellata are included. In fact, it seems that in Dinoflagellata the replacement of the plastid resulting from secondary endosymbiosis - the ancestral one in this group - by other plastids are very frequent events. This could be facilitated by the fact that Dinoflagellata are able to feed by myzocytosis (Figure 41), allowing plastids to be taken from endocytic vesicles directly in the cytoplasm of prey. In these flagellates, there are at least seven types of plastids and many other “photosynthetic endosymbionts” captured by kleptoplasty. Type 1 plastid with peridinin would therefore have been replaced repeatedly by others containing fuxoxanthin from diatoms (type 7), with 19-hexanoyloxyfucoxanthin from Haptophyta algae (types 2 and 4 resulting from two separate events), phycobilins probably coming from Cryptophyta algae (types 3 and 6 also resulting from two events) or with chlorophyll b then probably originating from Viridiplantae Prasinophyta (type 5)! These replacements are often referred to as tertiary endosymbioses, although not all of them are true endosymbiosis events involving algae from secondary endosymbiosis. For example, Amphidium latum or Gymnodinium acidotum capture Cryptophyta which they are able to store without digesting them. Cryptophyta largely retain their morphology but are not properly transmitted to the offspring, so these Dinoflagellata must make new endocytoses to perpetuate the association. Podolampas bipes and Podolampas reticulata contain Dictyophyta algae (a division of Ochrophyta) which is efficiently partitioned and transmitted during divisions to daughter cells. Others, such as Durinskia baltica or Kryptoperidinium foliaceum, have captured diatoms. Algae have lost much of their ultrastructure, but retain their nucleus and mitochondria. These algae are faithfully transmitted during the divisions and are therefore already very involved in the association. They therefore reproduce a new process of endosymbiosis and as they originate from a secondary endosymbiosis, it seems that in this case we are dealing with real tertiary endosymbiosis thus giving a new type of plastid (type 7). In fact, this type of plastid is surrounded by five membranes! In Karenia brevis and Karlodinium veneficum, Cryptophyta also resulting from a secondary endosymbiosis, the symbionts are an integral part of the cell because they are very modified and no longer have a cytoplasm. In this case, it is probably a plastid replacement by kleptoplasty because there appear to be less than four membranes around the plastid (their number is not clearly established, see also Box 7). The record appears to be in Dinophysis mitra, in whom more than 100 types of plastids have been detected by sequencing. Plastids from Haptophyta (type 4) seems to predominate because it is detected in 80% of cases, but sequences from Viridiplantae or Ochrophyta algae from various subgroups have been obtained. While kleptoplasty with Dinophysis mitra is no longer in doubt, it is unclear whether plastids are stably present and transmitted to the offspring. In contrast to these plastid gains, some appear to have been lost independently at least twice, in Crypthecodinium cohnii and in Oxyrrhis marina. In both cases, genes encoding plastid proteins are present in the nuclear genome, clearly demonstrating a loss in these species which have become phagotrophs again. Note that we will see in the next chapter that these Dinoflagellata often establish endosymbioses with other eukaryotes! These are the famous zooxanthellae that allow the survival of coral reefs.
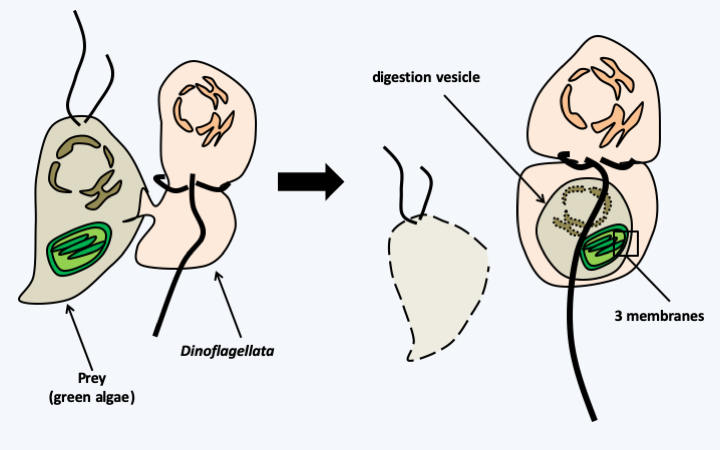
Figure 41.
Myzocytosis. Myzocytosis is a mode of nutrition in which the cytoplasm of the prey is directly 'pumped' into an endocytic vesicle. It can lead to kleptoplasty if the plastids persist in the endocytic vesicle. If the prey is an algae from primary endosymbiosis, the plastid is surrounded by three membranes. Plastids surrounded by three membranes are present in Dinoflagellata and Euglenida, groups which are able to feed themselves by myzocytosis.Box 8. Nucleomorphs
A nucleomorph is the remnant of the nucleus of an endosymbiotic eukaryotic alga whose integration is such that the alga is now considered a plastid (the plastid receives many proteins encoded by the host nucleus). Currently, it does not appear that the nucleomorph is involved in anything other than the biosynthesis of proteins destined for the plastid and it is considered to be a disappearing nucleus rather than an organelle. There are two groups of organisms in which nucleomorphs have been found: Chlorarachniophyta and Cryptophyta.
These two types of algae have, despite the presence of nucleomorphs, completely different origins. The Cryptophyta are related to the Haptophyta algae, while the Chlorarachniophyta are Rhizaria (see Figure 33). In the Cryptophyta, the plastid is derived from a Rhodophyta and its outermost membrane is continuous with that of the reticulum, while it is not in the Chlorarachniophyta, in which the plastid is derived from a Viridiplantae. The different characteristics of plastids mirror those of each of the algae that cause endosymbiosis. For example, the Cryptophyta store carbon reserves in the form of starch grains in the periplastid space, which is the space which contains the nucleomorph, therefore corresponding to the old cytoplasm of the endosymbiotic Rhodophyta. This space corresponds to where the reserves were stored in the Rhodophyta. Carbon reserves are stored in the cytoplasm in the form of β-glucans in Chlorarachniophyta. The storage location was therefore relocated and the reserve polymer changed in Chlorarachniophyta, because Viridiplantae store starch inside the plastids.
Nucleomorphs contain the 'smallest known eukaryotic genomes' as they measure around 500 kb. Chromosomes are linear and resemble chromosomes in eukaryotes in their overall structure. It is peculiar to see that there are three chromosomes in all the cases analyzed so far. However, the telomeric repeats of the chromosomes in Cryptophyta are variable and different from those of the usual telomeres (see table below). The percentage of the AT bases is very high. This base composition is also a characteristic of the plastid and mitochondrial genomes, and we do not yet know the reason. Obviously the same evolutionary pressures as those acting on organelles are exerted on nucleomorphs.
| Chlorarachniophyta | Cryptophyta | |
|---|---|---|
| Size of the haploid genome | 380–610 kb | 480–845 kb |
| Number of genes | 280-610 | 470-480 |
| GC content | 30-35% | 35-40% |
| Telomeres | [TCTAGGG]n | [(AG)7AAG6A]n/[(GA)17]4-7 |
| Operons | Present | Absent |
| Overlapping genes | Present | Absent |
| Introns of nuclear type | Numerous | Rare or absent |
The genomes of nucleomorphs have also been compacted by other mechanisms. The intergenic regions have been reduced and in some cases the genes overlap; in other cases, several ORFs are found on the same RNA and therefore constitute an operon. Note however that these operons have nothing to do with prokaryotic operons and we do not know how the proteins are expressed from the nucleomorph. Around the nucleomorph, there is a small cytoplasm containing ribosomes: these are encoded by the nucleomorph because in both groups there are rRNA genes encoding ribosomal proteins. Protein size is reduced especially in Cryptophyta. The nucleomorphs of Cryptophyta contain few introns spliced by the splicesome like the Rhodophyta genomes from which they derive, and that of Hemiselmis andersenii does not even have any! On the other hand, the nucleomorphs of Chlorarachniophyta contain a lot of introns, like the genomes of the Viridiplantae from which they derive. On the other hand, these introns are of very small size and represent, along with a few introns of Paramecium ciliates, the smallest introns known to be spliced. This poses an interesting problem, relevant to the evolution of introns. Clearly the selection pressure here is not the loss of introns but the decrease in their size: why?
There is no doubt that the analysis of these nucleomorphs, in particular their mode of expression, will reveal many interesting phenomena!
Plastids from secondary endosymbiosis have undergone the same changes as mitochondria and plastids from primary endosymbiosis: independent and frequent transfers of genes to the nucleus, modifications of the structure of genomes which, like in mitochondria, can take a very strange form in certain Dinoflagellata (Figure 42). There is even an early modification of the genetic code in the Apicomplexan Neospora caninum. Indeed, the rpoB gene of this species, encoding one of the subunits of RNA polymerase, contains three UGA stop codons in phase. These are decoded into tryptophan, possibly because the recognition of the UGA stop codon by the ribosome is impaired (the rRNA sequence of the small subunit is compatible with this pattern).
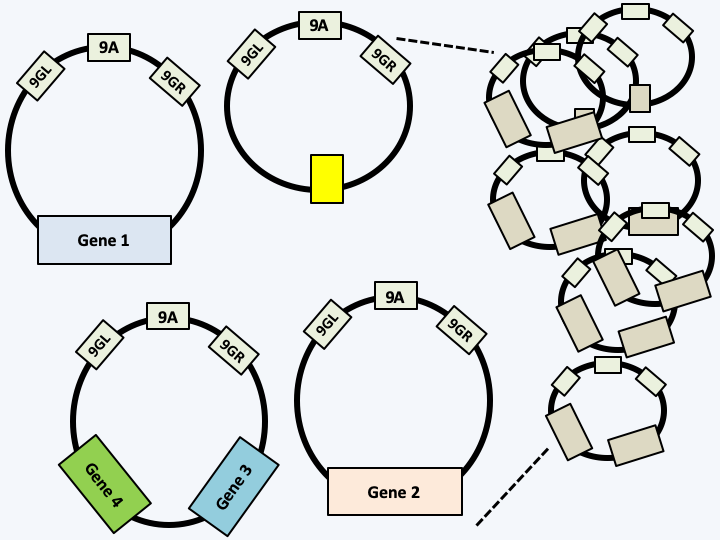
Figure 42.
Structure of the plastid DNA of Dinoflagellata possessing peridinin. The genome is composed of circular molecules measuring from 2 to 3 kb and comprising a common region with 3 very conserved sequences (9GL, 9A and 9GR) which are probably the origins of replication and transcription. Circles can encode one or two genes or none. The coding capacity is reduced because only about fifteen proteins are encoded by the genome of the plastid; the other genes have migrated into the nucleus. Partial fragmentation of the plastid genome is also observed in Karlodinium veneficum whose plastid originates from a Haptophyta.Successive endosymbioses, whether primary, secondary or others, have been accompanied by loss and modification of pigments explaining the diversity observed today. For example, all chlorophylls have an almost identical structure and diverge by a few groups and/or unsaturations which can evolve repeatedly and independently. Table 5 summarizes the origin and diversity of plastids in the different photosynthetic lineages of eukaryotes. So it seems that the chaotic evolution of plastids obscures rather than illuminates the evolution of eukaryotes. Be that as it may, primary endosymbiosis, which directly or indirectly gave rise to the majority of plastids present in eukaryotes, is along with mitochondrial endosymbiosis a major event in the history of eukaryotes. It allowed certain eukaryotes to feed themselves by carrying out oxygenic photosynthesis (Figure 43) and allowing eukaryotes to play the most important role in fixing atmospheric carbon.
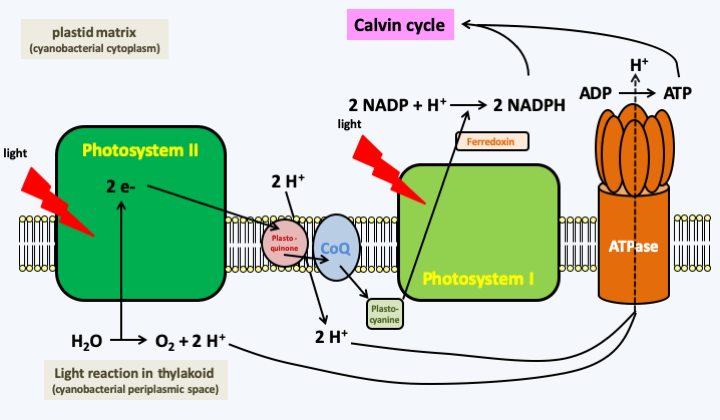
Figure 43.
Oxygenic photosynthesis. During photosynthesis, a water molecule is broken down into oxygen and a proton to recover two electrons which first travel through photosystem II and the other factors that act between the photosystems: plastoquinone, coenzyme-Q and plastocyanin. A second photon will be used to reenergize the electrons in photosystem I and allow transfer to NADP to produce NADPH. The electron travel is coupled with a proton transfer increasing the proton gradient between the plastid matrix and the thylakoid lumen which is topologically equivalent to the inter-membrane space. The protons then pass back through the matrix via an ATP synthetase, allowing the synthesis of ATP. The NADPH and ATP produced are then used to fix CO2 and synthesize sugars in a reaction that does not require light: the Calvin cycle. The different pigments allow better capture of photons and the transfer of their energy to photosystems.In bacteria, besides oxygenic photosynthesis, there are two other types of anoxygenic photosynthesis, that is, photosynthesis which does not produce oxygen. The first also uses multiprotein photosystems containing chlorophyll and is coupled to a Calvin cycle which allows CO2 to be fixed. This type of photosynthesis is restricted to eubacteria and does not seem to occur in eukaryotes or in archaea. The other type of photosynthesis (Figure 44) uses specific proteins, bacteriorhodopsins, which are related to the light-catching proteins in our retina. These proteins change their conformation by capturing a photon, which allows the translocation of a proton across the membrane where they are embedded. As in the case of classical photosynthesis, the proton gradient thus created can be used to synthesize ATP. Although this type of photosynthesis is not coupled with a Calvin cycle, ATP can be used as a source of energy by the cell. This mechanism is found in various prokaryotes and especially in archaea of the Halobacteria group. Rhodopsins that can act as a proton pump have been described in various eukaryotes, including fungi. These proteins could be involved in pH homeostasis or help generate a proton gradient for the facilitated transport of solutes. However, in some plastid-lacking Dinoflagellata, including Oxyrrhis marina, horizontal transfer of a bacteriorhodopsin gene has occurred from a eubacterium related to Proteobacteria. Dinoflagellata have not yet been shown to use these proteins to make ATP. But the gene, which is nuclear in Oxyrrhis marina, is highly expressed and the protein is located in an intracellular compartment, still unidentified, but which is not a plastid because the precursor does not have a signal peptide to go to a plastid. It could be the vacuole. The protein could then allow the synthesis of ATP via vacuolar ATPase, or else avoid the consumption of ATP to acidify the vacuole by using light, not ATP, as the energy source. Regardless, it appears that expression of the gene encoding bacteriorhodopsin is induced by light in Oxyrrhis marina and its survival is greater in the presence than in the absence of light when deprived of prey.
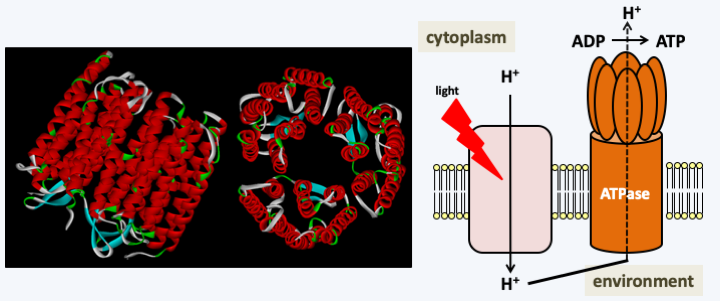
Figure 44.
Photosynthesis with rhodopsin. Bacteriorhodopsin (left) is a membrane protein that is able to change conformation when its co-factor, retinal, picks up a photon. This change in conformation is coupled with the transport of a proton across the membrane. The proton gradient thus created can be used for facilitated transport of solute or for the synthesis of ATP via a membrane ATPase. ATP production does not appear to be coupled with a CO2 binding mechanism and the production of sugars. This type of energy production from light energy is common in Halobacteria archaea, but may also occur in some Dinoflagellata. </figcaption> </figure>Endosymbiosis of the mitochondria and plastids have been permitted because eukaryotes have the ability to phagocytize whole cells. It is therefore not surprising that endosymbioses have played and continue to play essential roles in the evolution of eukaryotes. The next section will examine how these symbioses, whether intracellular or otherwise, come together and show their diversity in eukaryotes.
Back to chapter index